Amino acid sequences and evolutionary relationships answer key – Unveiling the mysteries of evolutionary relationships, the study of amino acid sequences and their profound implications takes center stage. As the building blocks of proteins, these sequences hold a wealth of information, allowing scientists to trace the intricate threads that connect species throughout the vast tapestry of life.
This discourse delves into the comparative analysis of amino acid sequences, revealing how they illuminate evolutionary history. We explore the techniques of sequence alignment and phylogenetic tree construction, unlocking the secrets of homologous regions and genetic divergence. Furthermore, the concept of the molecular clock provides a temporal framework, enabling us to estimate evolutionary timelines and unravel the grand narrative of life’s journey.
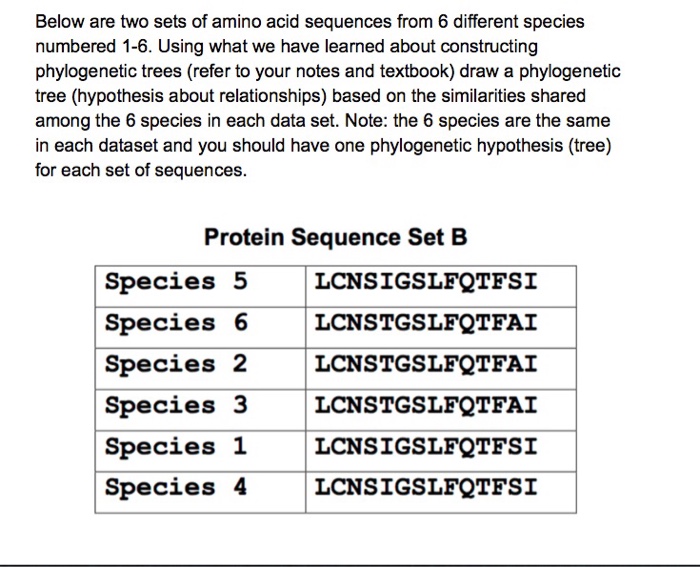
Amino Acid Sequence Comparison
Amino acid sequences provide valuable insights into the evolutionary relationships between different organisms. By comparing the sequences of amino acids in proteins, scientists can determine the degree of similarity and divergence between species, providing evidence for common ancestry and the evolutionary history of life.
Methods used to compare amino acid sequences include:
- Sequence alignment: Aligning sequences to identify regions of similarity and difference.
- Phylogenetic tree construction: Using aligned sequences to build trees that represent evolutionary relationships.
The length and similarity of amino acid sequences play a crucial role in determining evolutionary relationships. Longer sequences provide more data for comparison, while higher similarity suggests a closer evolutionary relationship.
Sequence Alignment
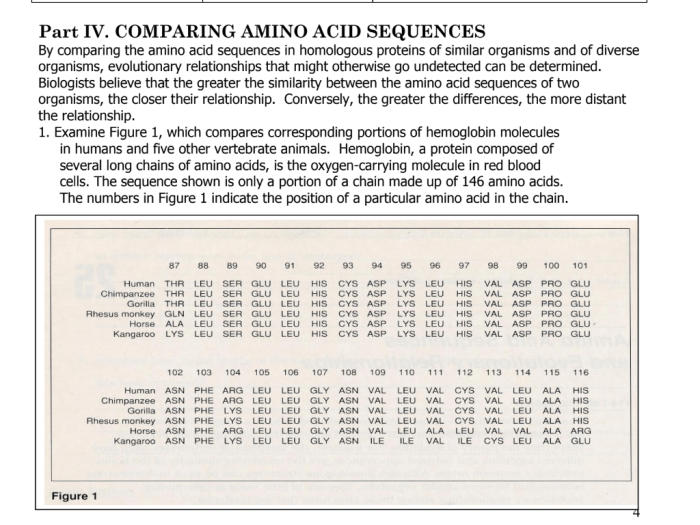
Sequence alignment is a process of arranging two or more sequences to identify regions of similarity and difference. This helps identify homologous regions, which are sequences that have descended from a common ancestral sequence.
An example of a sequence alignment between two different protein sequences:
| Sequence 1 | A-G-T-C-D-E-F-G-H-I-J |
|---|---|
| Sequence 2 | A-G-T-C-D-E-F-X-H-I-J |
In this alignment, the “X” represents a gap introduced to maximize sequence similarity. Gaps indicate regions where insertions or deletions have occurred during evolution.
Phylogenetic Tree Construction, Amino acid sequences and evolutionary relationships answer key
Phylogenetic trees are diagrams that represent evolutionary relationships based on amino acid sequences. They depict the branching patterns of different species, with the root of the tree representing the common ancestor and the branches representing evolutionary lineages.
Methods used to construct phylogenetic trees include:
- Neighbor-joining: A method that uses distance measures to group similar sequences together.
- Maximum parsimony: A method that seeks to find the tree with the fewest evolutionary changes.
An example of a phylogenetic tree based on amino acid sequences:

Molecular Clock
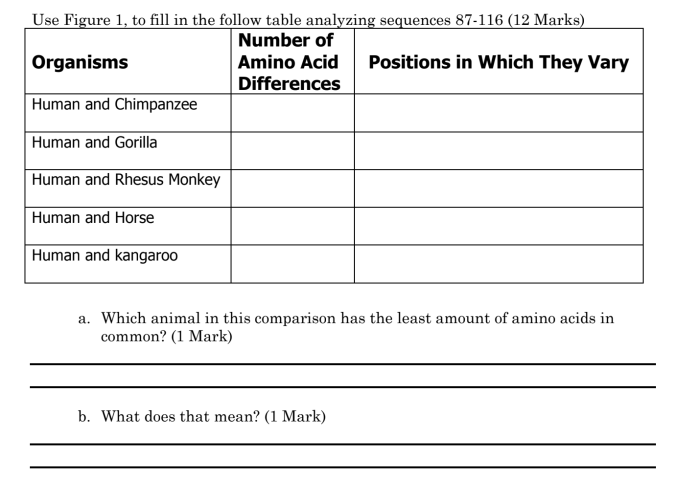
The molecular clock is a concept that suggests that the rate of molecular evolution, such as amino acid substitutions, is relatively constant over time. This allows scientists to estimate the divergence times between species by comparing the number of amino acid differences between their sequences.
Assumptions and limitations of the molecular clock include:
- The rate of evolution is constant across all lineages.
- The rate of evolution is not influenced by environmental factors.
Despite these limitations, the molecular clock has been used to study evolutionary relationships in a variety of organisms, including humans and other primates.
Essential Questionnaire: Amino Acid Sequences And Evolutionary Relationships Answer Key
What is the significance of sequence length and similarity in determining evolutionary relationships?
Sequence length and similarity play a crucial role in determining evolutionary relationships. Longer sequences provide more data points for comparison, increasing the accuracy of analysis. Higher sequence similarity suggests a closer evolutionary relationship, as it indicates a greater degree of shared genetic heritage.
How does the molecular clock contribute to the study of evolutionary relationships?
The molecular clock assumes a constant rate of genetic change over time. By comparing the genetic differences between species, scientists can estimate the time since their divergence from a common ancestor. This provides a temporal framework for understanding evolutionary relationships and the pace of genetic evolution.
What is the process of sequence alignment and its role in identifying homologous regions?
Sequence alignment involves arranging two or more sequences side-by-side to identify regions of similarity. Homologous regions represent conserved sequences that have been inherited from a common ancestor. Identifying homologous regions is crucial for inferring evolutionary relationships, as it allows researchers to trace the descent of genetic material across species.